Kröger Group
Welcome to the homepage of Kröger Group at the Technical University of Dresden, Germany
 © Kroeger
© Kroeger
Biomineralization, Bioadhesion and Bio-enabled Materials Synthesis
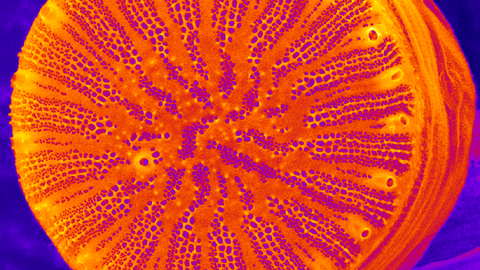
The research of the Kröger group focuses on diatoms, which are a large group of eukaryotic microalgae. We apply biochemical, molecular genetic, and cell biological methods to investigate two remarkable biological phenomena exhibited by diatoms: silica biomineralization and underwater adhesion.