 © TUD/Philip Gröger
© TUD/Philip Gröger
Molecular Biophysics: Conformational Dynamics of Biomolecules
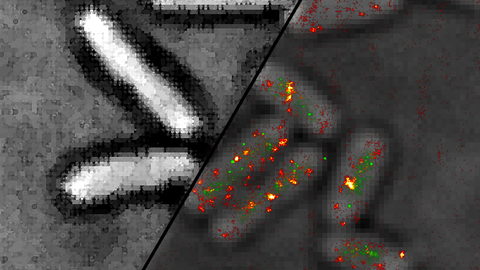
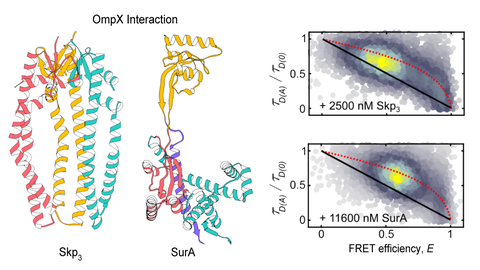
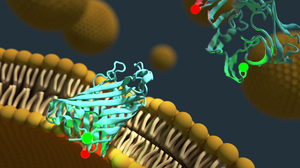
We are interested in conformational dynamics of biomolecules. In particular, we study (membrane) protein folding, protein-DNA interactions during DNA replication and recombination and regulatory DNA and RNA conformations.